|

|
Math 241 - Maple Workshop
Fall Semester, 2003
Vectors and Graphics
|
|
|
|
© 2001, All Rights Reserved, SDSU &
Joseph M. Mahaffy
San Diego State University -- This page last updated 13-Oct-03
|
|
Vector and Graphics
This session will examine several graphics
routines with vector Calculus, then show how Maple can be applied to differential
equations. Data is fit to
a cubic polynomial, then graphed.
3-D surfaces
are plotted, then the volume
and surface integrals
are analyzed. The Divergence
and Stokes' Theorems are visualized, then
special functions, including gradient
and curl
are applied. We include the LaTeX
command to show how to generate latex code for good document presentation.
The graphics ends with animations of Fourier
series and a parametrized
3-D surface.
As usual, there is a hyperlink to the Maple Worksheet
for this lecture.
Vectors
We begin with some basic vector operations.
> a := [2, -1, 5]; b := [1,
3, -4]; c := Vector[row](3,-2);
![a := [2, -1, 5]](images/vect1.gif)
![b := [1, 3, -4]](images/vect2.gif)
![c := _rtable[1293560]](images/vect3.gif)
Adding vectors is easy, but different forms
may require the evalm (matrix evaluation) command.
> a+b;evalm(a+b-c);
![[3, 2, 1]](images/vect4.gif)
![vector([5, 4, 3])](images/vect5.gif)
Many of the operations with vectors require
one of the Maple special packages, linalg or LinearAlgebra.
> with(linalg):
Below we show the dot or inner product and
the cross product.
> dotprod(a,b); innerprod(a,c);
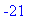
> crossprod(a,b);
![vector([-11, 13, 7])](images/vect7.gif)
Next we enter a vector function, then take
the divergence and curl of this function.
> F := (x,y,z) -> [x^2*cos(y*z),-x^2*z*exp(y^2),ln(x)/(z*y)];
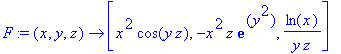
> diverge(F(x,y,z),[x,y,z]);
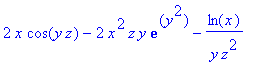
> curl(F(x,y,z),[x,y,z]);
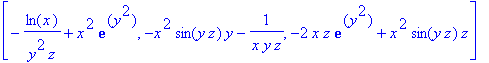
Graphics
Maple's graphics capabilities cover a
variety areas using several special packages. These require special
tools and some are shown below.
Fitting Data
Suppose we want to fit data to a cubic model
and visualize both the data and the model. Our example comes from the
oxygen consumption of the "kissing bug,"
Triatoma phyllosoma
, during
the fourth instar stage (Data from Boyd Collier). This bug carries
chagas disease. The time data (in hours) is stored as td, while the
oxygen consumption is stored as yd.
> td := [1, 2, 3, 4, 5,
6, 7, 8, 9, 10, 11, 12, 13, 14, 15]: yd := [116.6, 120.1,
114.9, 129.9, 116.5, 107.7, 99.0, 104.0, 100.7, 87.5, 82.7, 53.8,
54.0, 72.4, 81.1]:
We will use the statistics package (for
least squares), the statistics plotting package (for scatterplot),
and the plotting package (for displaying two types of
graphs).
> with(stats):
with(stats[statplots]): with(plots):
Graph the data with circles for points, then
store this graph for later. First view the graph, then store the
picture.
>
plots[display](scatterplot(td,yd),symbol=circle);
![[Maple Plot]](images/session31.gif)
> Data :=
plots[display](scatterplot(td,yd),symbol=circle):
Create a general cubic
model.
> model := a*t^3 + b*t^2 +
c*t + d;
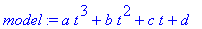
Find the least squares best fit of the cubic
model to the data.
> eqn :=
fit[leastsquare[[t,y], y=model,
{a,b,c,d}]]([td, yd]);
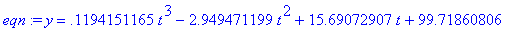
Make
y into a
function depending on t
.
> y :=
unapply(rhs(eqn),t);
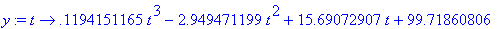
Store the graph of the
model, then display it with the graph of the data.
> Y :=
plot(y(t),t=0..15):
>
display({Data,Y});
![[Maple Plot]](images/session35.gif)
Select individual coefficients from the
model.
> b :=
coeff(y(t),t,2);
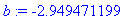
Volume and Surface Area
Maple provides a good tool for visualizing
the 3-D surfaces from vector Calculus. It can be readily used to find
the surface area and volume of these 3-D objects. Consider the
following paraboloid.
S =
{( x
, y
, z
)|
z = 4 -
4 x2 -
y2,
z
>
0}
To visualize this surface in Maple, we need
to parameterize the x
, y
, and
z
components into 2 parameters. I want the graph centered on the origin
(axes = NORMAL) and the picture unscaled (scaling =
CONSTRAINED).
> r :=
[u*cos(v)/2,u*sin(v),4-u^2];
![r := [1/2*u*cos(v), u*sin(v), 4-u^2]](images/session37.gif)
>
plot3d([r[1],r[2],r[3]],u=0..2,v=0..2*Pi,axes
= NORMAL, labels = [x,y,z], scaling = CONSTRAINED,
orientation = [40,70]);
![[Maple Plot]](images/session38.gif)
We find the volume under
this surface (using symmetry).
>
4*Int(Int(4-4*x^2-y^2,y=0..sqrt(4-4*x^2)),x=0..1);
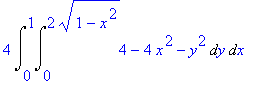
The formula can be inserted into a
LaTeX document by letting Maple generate the
code.
> latex(%);
4\,\int _{0}^{1}\!\int _{0}^{2\,\sqrt {1-{x}^{2}}}\!4-4\,{x}^{2}-{y}^{
2}{dy}\,{dx}
Below we compute the double
integral.
>
4*int(int(4-4*x^2-y^2,y=0..sqrt(4-4*x^2)),x=0..1);
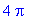
We find the surface area
of this surface (again using symmetry). A reminder that if
z =
f
( x
, y
), then the formula for computing the area of
the surface is
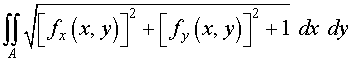
> f := (x,y) -> 4 - 4*x^2
- y^2;
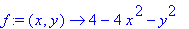
>
4*Int(Int(sqrt((diff(f(x,y),x))^2+(diff(f(x,y),y))^2+1),y=0..sqrt(4-4*x^2)),x=0..1);
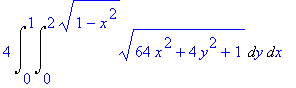
>
4*int(int(sqrt((diff(f(x,y),x))^2+(diff(f(x,y),y))^2+1),y=0..sqrt(4-4*x^2)),x=0..1);
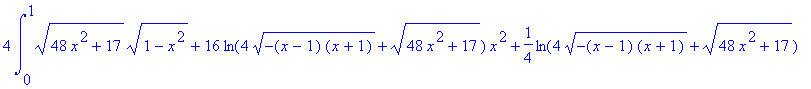
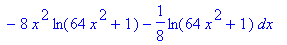
> evalf(%);
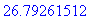
An alternate method for computing surface
integrals uses the following formula
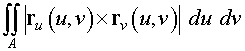
>
with(linalg):
We created the parametrized surface by the
function r
( u
, v
) given above. By taking the crossproduct of
the partial derivatives of the parametrized surface function, we
obtain the outward normal to the surface.
> ruxrv :=
crossprod(diff(r,u),diff(r,v));
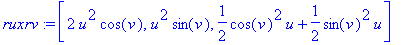
> absruxrv :=
sqrt(ruxrv[1]^2 + ruxrv[2]^2 +
ruxrv[3]^2);
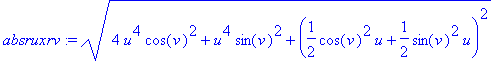
The area of the surface is the integral over
the range of the parameters of the magnitude of the outward normal.
One could use the norm function, but Maple has difficulty integrating
quantities with absolute values.
> int(int(absruxrv, u =
0..2),v = 0..2*Pi);

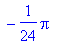
Often when the integral cannot be evaluated
exactly, the evalf function allows numerical solution of the
integral.
> evalf(%);
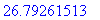
Gauss' and Stokes'
Theorems
Two of the major theorems used for fluid
flow are the Divergence or Gauss' Theorem and Stokes' Theorem. The
Divergence Theorem provides an means of determining the flow through
a bounded region in 3-space. Stokes' Theorem gives information on the
rotation of a fluid.
Let us consider these
theorems for a fluid flowing through a hemisphere. We begin by
visualizing the fluid flow and the hemisphere.
>
with(plots):
View the fluid flow, using Maple's
fieldplot.
> F := [x^2*y, y*z,
2*z-1];
![F := [x^2*y, y*z, 2*z-1]](images/session321.gif)
>
fieldplot3d(F,x=-4..4,y=-4..4,z=-1..4,grid=[8,8,5],axes
= NORMAL, thickness = 2);
Flow :=
fieldplot3d(F,x=-4..4,y=-4..4,z=-1..4,grid=[8,8,5],axes =
NORMAL, thickness = 2):
![[Maple Plot]](images/session322.gif)
Graph the hemisphere, which is the surface
bounding the region for applying the Divergence and Stokes'
Theorem.
> r :=
[3*cos(u)*cos(v),3*sin(u)*cos(v),3*sin(v)];
![r := [3*cos(u)*cos(v), 3*sin(u)*cos(v), 3*sin(v)]](images/session323.gif)
>
plot3d([r[1],r[2],r[3]],u=0..2*Pi,v=0..Pi/2,axes
= NORMAL, labels = [x,y,z], orientation = [40,70],
style = WIREFRAME, color = BLACK);
Hemi :=
plot3d([r[1],r[2],r[3]],u=0..2*Pi,v=0..Pi/2,axes
= NORMAL, labels = [x,y,z], scaling = CONSTRAINED,
orientation = [40,70], style = WIREFRAME, color =
BLACK):
![[Maple Plot]](images/session324.gif)
>
display3d({Flow,Hemi});
![[Maple Plot]](images/session325.gif)
The Divergence Theorem is given
by
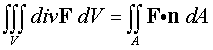
The easiest way to solve the second integral is to parametrize
the surface A, as A = r(u,v), then
use this parameterization to find the normal to the surface,
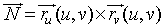
The integral becomes
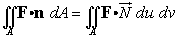
>
with(linalg):
Here we compute the
divergence of the vector flow field, then integrate
this quantity over the volume.
> divF :=
diverge(F,[x,y,z]);
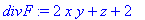
> Int(Int(Int(divF,z =
0..sqrt(9-x^2-y^2)),y = -sqrt(9-x^2)..sqrt(9-x^2)),x =
-3..3);
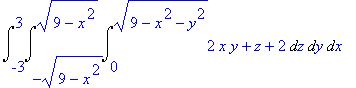
> int(int(int(divF,z =
0..sqrt(9-x^2-y^2)),y = -sqrt(9-x^2)..sqrt(9-x^2)),x =
-3..3);
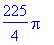
> evalf(%);
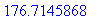
The above integral is more easily written in
spherical coordinates and given by
>
Int(Int(Int((2*rho^2*cos(theta)*sin(theta)*sin(phi)^2
+ rho*cos(phi) + 2)*rho^2*sin(phi),rho = 0..3),theta = 0..2*Pi),phi =
0..Pi/2);
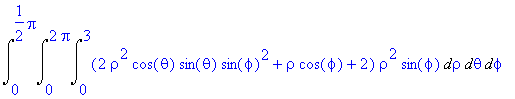
>
int(int(int((2*rho^2*cos(theta)*sin(theta)*sin(phi)^2
+ rho*cos(phi) + 2)*rho^2*sin(phi),rho = 0..3),theta = 0..2*Pi),phi =
0..Pi/2);
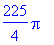
> evalf(%);
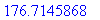
Stokes' Theorem measures
the circulation around any curve
C . If we
consider the curve where the hemisphere intersects the
xy -plane
with the flow F
given above, then Stokes' Theorem states
that
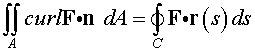
The integral on the right is most easily solved using the technique
above for the Divergence Theorem. Parametrize the surface A = r(u,v),
then use this parameterization to find the normal to the surface,
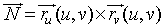
The double integral becomes the same as above with curl F
replacing F.
Here we compute the curl of the vector
field, then evaluate the double integral over the circle in the
xy
-plane beneath the hemisphere.
> curlF :=
curl(F,[x,y,z]);
![curlF := vector([-y, 0, -x^2])](images/session333.gif)
> curlFn :=
dotprod(curlF,[0,0,1]);
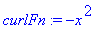
> Int(Int(curlFn,y =
-sqrt(9-x^2)..sqrt(9-x^2)),x = -3..3);
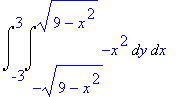
> int(int(curlFn,y =
-sqrt(9-x^2)..sqrt(9-x^2)),x = -3..3);
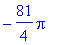
In polar coordinates...
>
int(int(-R^3*cos(theta)^2,R = 0..3),theta =
0..2*Pi);
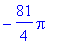
Animations
Maple has programs that allow visualization
of 2 and 3-D graphics through animations.
We begin with a repeat of our
Fourier series expansion from the previous
session. However, this time, the evolution of the partial sums of the
Fourier series is viewed through animation, which allows one to
easily see how the Fourier series is converging. The animation
program requires the plots package.
>
with(plots):
> bi :=
int(x*sin(i*Pi*x),x=-1..1);
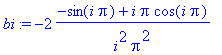
> S := n ->
sum(bi*sin(i*Pi*x), i = 1..n);
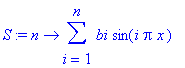
> animate(S(n),
x=-3..3,n=1..20,frames=20,numpoints=500, color =
blue);
![[Maple Plot]](images/session340.gif)
After invoking the animation, a series of
buttons appears on the menu bar. These allow the user to view the
movie of the graph evolving either forward or backward, cycle
through, or go frame by frame.
Below we simply took one of the
3-D examples from Maple's Help. In the 3-D case,
the default uses just 8 frames.
>
animate3d(cos(t*x)*sin(t*y),x=-Pi..Pi,
y=-Pi..Pi,t=1..2,color=cos(x*y));
![[Maple Plot]](images/session341.gif)
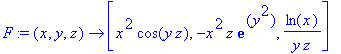
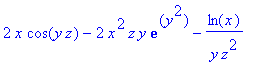
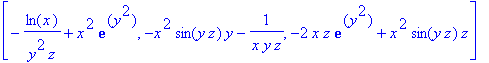
![[Maple Plot]](images/session31.gif)
![[Maple Plot]](images/session35.gif)
![[Maple Plot]](images/session38.gif)
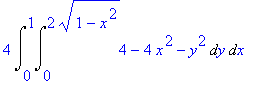
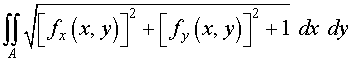
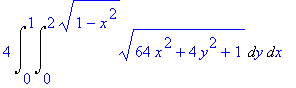
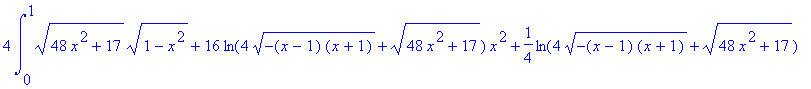
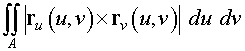
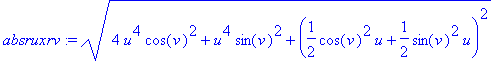
![[Maple Plot]](images/session322.gif)
![[Maple Plot]](images/session324.gif)
![[Maple Plot]](images/session325.gif)
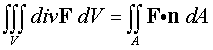
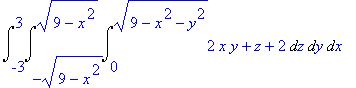
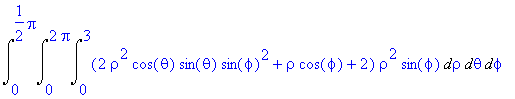
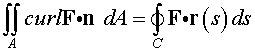
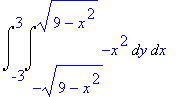
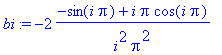
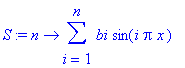
![[Maple Plot]](images/session340.gif)
![[Maple Plot]](images/session341.gif)